Authors: Alireza Omidi, Mads Harder Møller, Nawar Malhis, Jennifer M. Bui, Jörg Gsponer
Published in Proceedings of the National Academy of Sciences (PNAS), 2024
Abstract
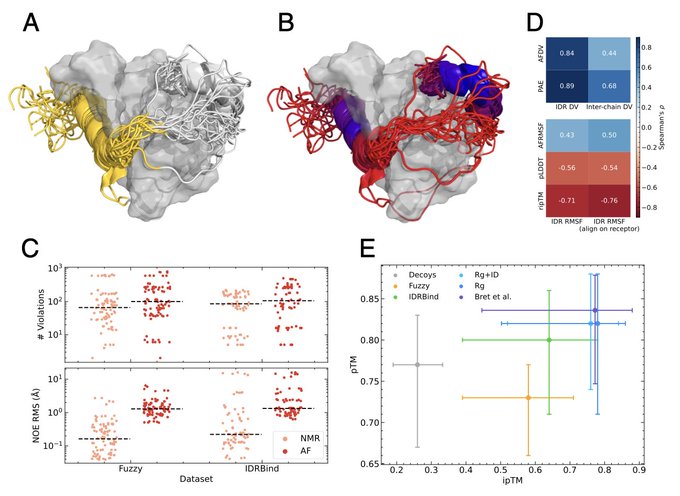
Interactions mediated by intrinsically disordered protein regions (IDRs) pose formidable challenges in structural characterization. IDRs are highly versatile, capable of adopting diverse structures and engagement modes. Motivated by recent strides in protein structure prediction, we embarked on exploring the extent to which AlphaFold-Multimer can faithfully reproduce the intricacies of interactions involving IDRs. To this end, we gathered multiple datasets covering the versatile spectrum of IDR binding modes and used them to probe AlphaFold-Multimer's prediction of IDR interactions and their dynamics. Our analyses revealed that AlphaFold-Multimer is not only capable of predicting various types of bound IDR structures with high success rate, but that distinguishing true interactions from decoys, and unreliable predictions from accurate ones is achievable by appropriate use of AlphaFold-Multimer's intrinsic scores.