Authors: Anastasia Litinetskaya, Maiia Schulman, Fabiola Curion, Artur Szalata, Alireza Omidi, Mohammad Lotfollahi, Fabian Theis
Published in Research in Computational Molecular Biology (RECOMB), 2025
Abstract
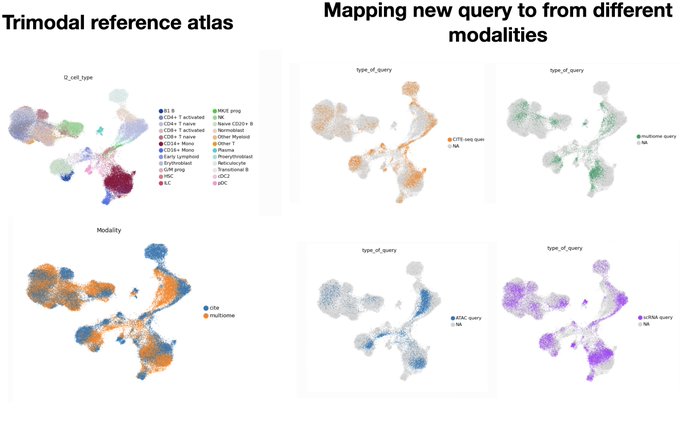
Constructing joint representations from multimodal single-cell datasets is crucial for understanding cellular heterogeneity and function. In this work, we demonstrate the product-of-experts VAE-based model, which offers a flexible, scalable solution for integrating multimodal data, allowing for the seamless mapping of both unimodal and multimodal queries onto a reference atlas. We evaluate how different strategies for combining modalities in the VAE framework impact query-to-reference mapping across diverse datasets, including CITE-seq and spatial metabolomics. We showcase our approach in a mosaic setting, integrating CITE-seq and multiome data to accurately map unimodal and multimodal queries into the joint latent space. We extend this to spatial data by integrating gene expression and metabolomics from paired Visium and MALDI-MSI slides, achieving a high correlation in metabolite predictions from spatial gene expression. Our results demonstrate that this VAE-based framework is scalable, robust, and easily applicable across multiple modalities, providing a powerful tool for data imputation, querying, and biological discovery.
Are you wondering how to integrate scRNAseq and scATAC-seq or combine it with a CITE-seq to have tiromodal single-cell reference atlas? How about mapping a new query data (e.g., a new scATAC or scRNA)? Check out our new Multigrate vignette: scarches.readthedocs.io/en/latest/mult…